| Team Members: | Annie Dai1, David Palensky2, Alex Piatski3, Kendall Queen4, and Gina Vockeroth5 |
| Graduate Research Assistant: | Teresa Lebair6 |
| Faculty Mentor: | Bradford E. Peercy6 |
| Client: | Margaret Watts7 and Arthur Sherman7 |
1Department of Mathematics, Northeastern University,
2Department of Mathematics, University of Maryland, College Park,
3Department of Computer and Information Science, University of Pennsylvania,
4Department of Computer Science and Electrical Engineering, University of Maryland, Baltimore County,
5Department of Mathematics and Statistics, Loyola University Chicago
6Department of Mathematics and Statistics, University of Maryland, Baltimore County,
7Laboratory of Biological Modeling, National Institutes of Health

Team 3, from left to right: Teresa Lebair, David Palensky, Alex Piatski, Gina Vockeroth, Bradford E. Peercy, Annie Dai, Kendall Queen.
About the Team
Our team, Annie Dai, David Palensky, Alex Piatski, Kendall Queen, and Gina Vockeroth, participated in the REU Site: Interdisciplinary Program in High Performance Computing located in the Department of Mathematics and Statistics at UMBC. Our project focused on modeling pancreatic islets, which consist of alpha, beta, and delta cells. Through the assistance of our faculty mentor, Dr. Bradford E. Peercy from Mathematics and Statistics Department at UMBC, our graduate research assistant, Teresa Lebair also from Mathematics and Statistics Department at UMBC, and our clients, Dr. Arthur Sherman and Dr. Margaret Watts from the Laboratory of Biological Modeling at NIH, our team was able to produce accurate simulations of how alpha, beta, and delta cells react with each other in a given space.
Problem Introduction
Diabetes is a pervasive, metabolic disease of elevated blood glucose levels. In order to understand this disease it is important to study the structure and function of units in the pancreas called islets of Langerhans. The cells that make up these islets are responsible for insulin regulation. Our mathematical model focuses on alpha, beta, and delta cells. We have adapted a Tri-Hormone Model into a multicellular computational islet to study 1) the effects of cell-type ratio dependence on secretion through paracrine interactions, 2) paracrine feedback on synchronizing cellular heterogeneity within cell-type, and 3) spatial distribution of cell-types and its effect on secretion.
Expanding the Tri-Hormone Model
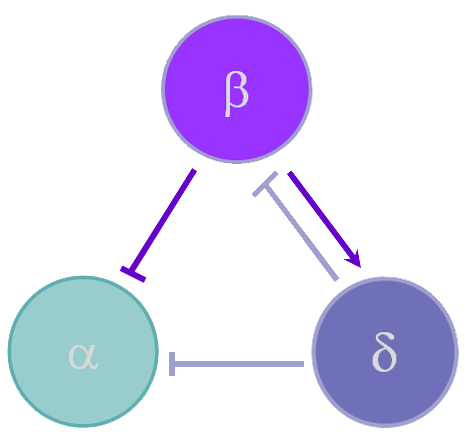
Figure 1Experimental Cases
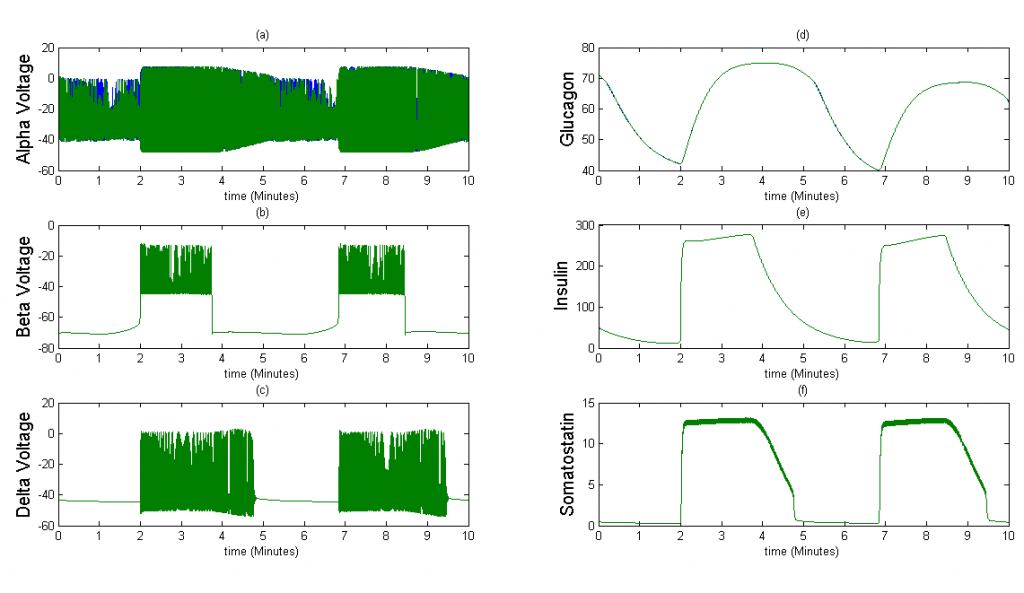
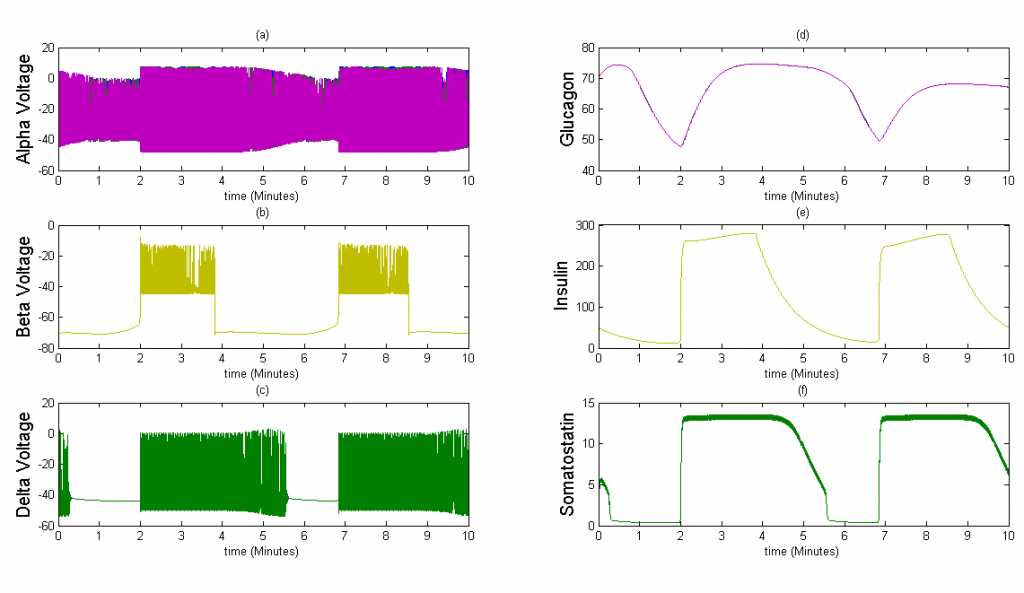
Once we were able to scale the Tri-Hormone model we ran simulations for two different 3x3x3 cubic islets. These islets are compartmental, which means that each cell detects the effects of another cell instantaneously. The first case represents an islet with 9 of each cell type organized in 3 sequential planes. The secretion from beta and delta cells is modeled as the sum divided by 9 cells, in order to simulate the average secretion from each cell. The second case is an accurate percentage representation of a mouse islet. This islet contains 5 alpha cells, 20 beta cells, and 2 delta cells. We also keep the secretion constant with the sum of insulin and somatostatin divided by 9 in case 1 to observe how changes in the distribution of cells affect islet behavior.
 |
 |
| Equal Number of cells (9: 9: 9) | Mouse Islet Representation (5: 20: 2) |
Figure 2
Spatial Islet Diffusion
The Tri-Hormone model, we replicated does not have the spatial diffusion of cell secretion. Each cell secretes into a shared space and each cell is affected by the same amount of secretion in this space. We extend our model with the spatial diffusion and now every cell is influenced by the amount of secretion in its space, we can also make a three-dimensional representation of how hormone secrete into space.
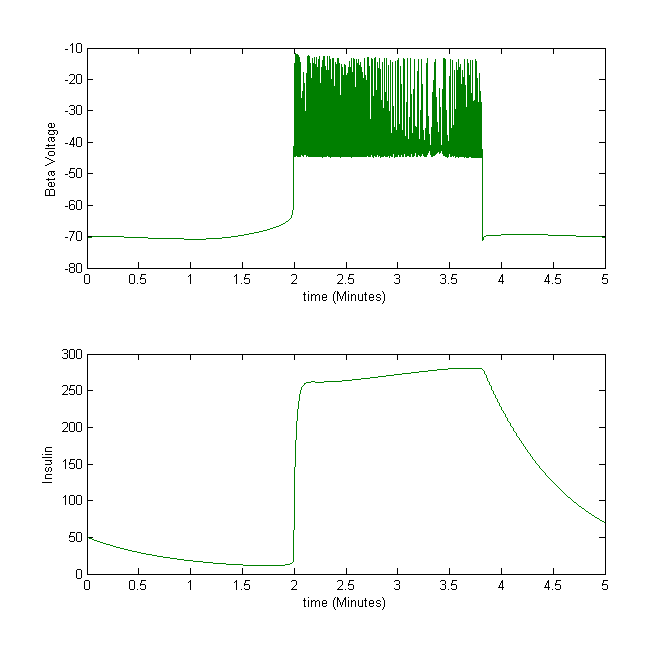
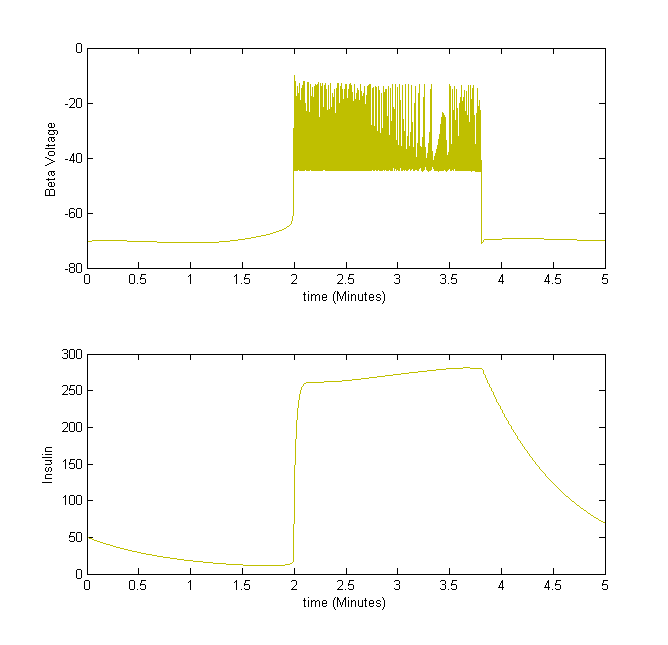
For example, we take the insulin secretion from beta cells. When the beta cell voltage is bursting, insulin is secreted.
 |
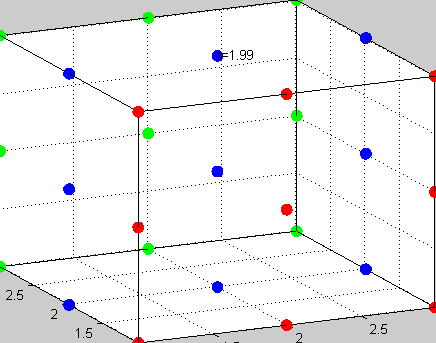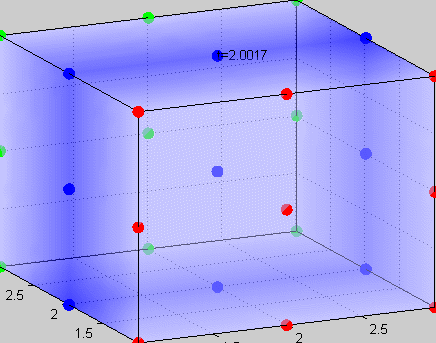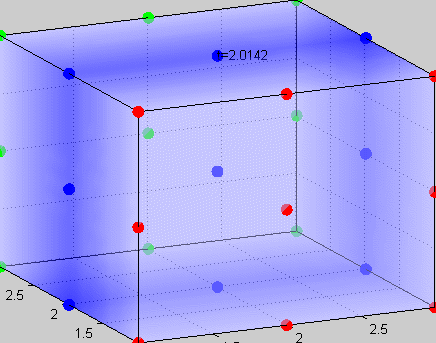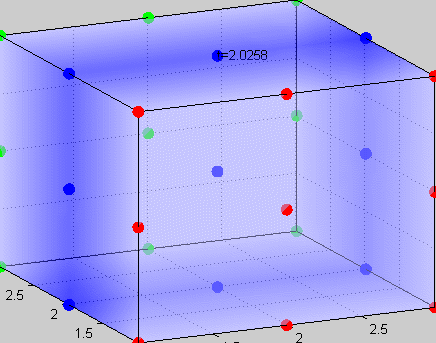 |
| 3a | 3b |
Figure 3The above graph has the distribution of islet cells by 9:9:9 (alpha:beta:delta). The left top figure is the beta voltage, we can see that it starts bursting at two minutes, the left bottom figure shows the amount of insulin secreted by each beta cell corresponding to time, and we can see that at two minutes, it rapidly increases. The right animated figure is the spatial representation of the insulin secretion around the time at two minutes. The dots represent the position of each cell in the space, red dots are alpha cells, blue dots are beta cells and green dots are delta cells. Since the amount of secretion is increasing too rapidly, from the animated figure, we can see that insulin secreted from beta cells fill out the entire space in less than a second.
 |
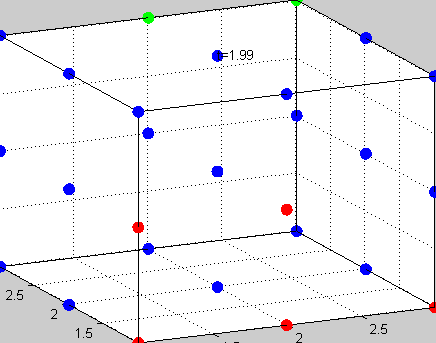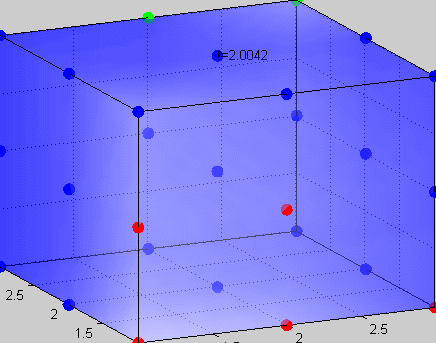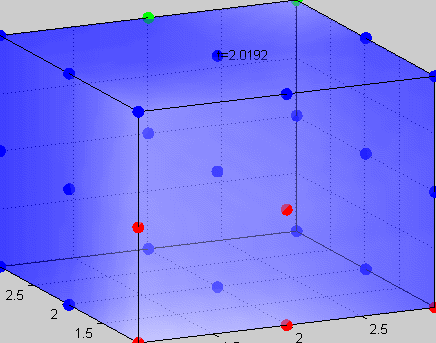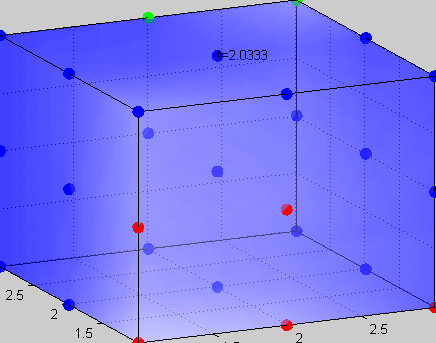 |
| 4a | 4b |
Figure 4The above graph has the cell type distribution of mouse-like islet: 5:20:2 (alpha:beta:delta). The voltage looks similar to the previous figure and the amount of insulin secreted by each beta cell is similar. However, since there are many more beta cells in this model, the spatial insulin diffusion is different; see the animated figure on the right.
Heterogeneity of Alpha Cells
In order to simulate the effects of paracrine coupling on the heterogeneity of alpha cells, we assigned two different GK(ATP) values (26.5*Dza+0.04,32*Dza+0.04) to the alpha cells in our islet. This GK(ATP) value is the conductance for the K(ATP) channel in the alpha cell, which varies depending on the amount of glucose in the blood. We simulate a high blood glucose level with our GK(ATP) = 26.5* Dza + 0.04. We ran tests with and without the effects of beta and delta cells felt on alpha cells to see if paracrine coupling can tame the heterogeneity of alpha cells. The blue bursts correspond to GK(ATP)=26.5*Dza + 0.04, the green bursts correspond to GK(ATP) = 32*Dza + 0.04.
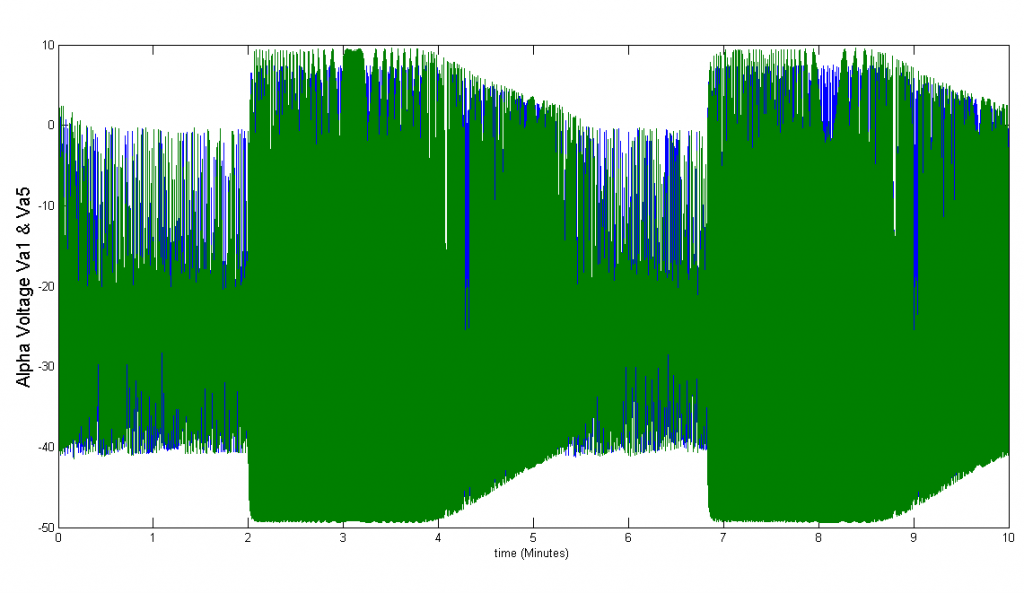
Figure 3
Conclusions
We were able to accurately scale up the Tri-Hormone model after translating XPP code to MATLAB. We were also able to successfully implement Beta cell and paracrine coupling in Matlab. We proved that paracrine coupling effects can,in fact, tame heterogeneity. Overall, our team was able to produce a multicellular, dynamic, discretized, mathematical model that can visually and computationally represent hormonal diffusion and cell interaction of a pancreatic islet of Langerhans.
Links
Annie Dai, David Palensky, Alex Piatski, Kendall Queen, Gina Vockeroth, Teresa Lebair, Bradford E. Peercy, Margaret Watts, and Arthur Sherman. Modeling the Building Blocks of the Pancreatic Islet: Connecting Alpha, Beta, and Delta Cells. Technical Report HPCF-2014-13, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, 2014. (HPCF machines used: maya.). Reprint in HPCF publications list
Poster presented at the Summer Undergraduate Research Fest (SURF)
Click here to view Team 1’s project
Click here to view Team 2’s project
Click here to view Team 4’s project
Click here to view Team 5’s project