| Team Members: | Nil Mistry1, Jordan Ramsey2, Benjamin Wiley3, and Jackie Yanchuck4 |
| Graduate Research Assistants: | Xuan Huang5 and Andrew Raim5 |
| Faculty Mentor: | Matthias K. Gobbert5 and Nagaraj K. Neerchal5 |
| Client: | Philip J. Farabaugh6, Christopher Mineo7, and David Mountain7 |
1Department of Mathematics and Statistics, University of Connecticut,
2Department of Computer Science and Electrical Engineering, University of Maryland, Baltimore County,
3Department of Mathematics and Statistics, University of New Mexico,
4Department of Mathematics, Seton Hill University,
5Department of Mathematics and Statistics, University of Maryland, Baltimore County,
6Department of Biological Sciences, University of Maryland, Baltimore County,
7Advanced Computing Systems Research Program

Team 3, from left to right: Jordan Ramsey, Nil Mistry, Benjamin Wiley, Jackie Yanchuck
About the Team
Our team consists of four undergraduates, Nil Mistry, Jordan Ramsey, Benjamin Wiley, and Jackie Yankchuk. Our group was interested in two projects that were presented to us and decided to do both of them. The first project is Contention of Communications in Switched Networks with Applications to Parallel Sorting. In this project, our research assistant was graduate student Xuan Huang, faculty mentor Dr. Matthias Gobbert, and clients Christopher Mineo and David Mountain with the Advanced Computing Systems Research Program. Our second project isClustering of Multidimensional Datasets with Applications to Spatial Distributions of Ribosomal Proteins with research assistants Xuan Huang and Andrew Raim, faculty mentors Matthias K. Gobbert and Nagaraj K. Neerchal, and client Philip Farabaugh of the UMBC Biology Department.
Contention of Communications in Switched Networks with Applications to Parallel Sorting
Contention of communications across a switched network that connects multiple compute nodes in a distributed-memory cluster may seriously degrade the performance of parallel code. This contention is maximized when communicating large blocks of data among all parallel processes simultaneously. This communication pattern arises in many important algorithms such as parallel sorting. InfiniBand interconnects are the most popular high-performance networks in computing clusters presently. We use the cluster tara in the UMBC High Performance Computing Facility (HPCF) with a quad-data rate InfiniBand network to provide a test case if the capacity of a switched network can be a limiting factor in algorithmic performance.
The distributed-memory cluster tara contains 82 compute nodes (arranged in two stacks of ‘pizza boxes’ in the racks), each with two quad-core Intel Nehalem X5550 processors (2.66GHz, 8192kB cache) and 24GB of local memory. All nodes on tara are connected by a high-performance quad-data rate InfiniBand network. Each node connects by cable (the red fiber-optic cables in Figure 1a) to one port in an 18-port InfiniBand leaf module, see Figure 1b. The central InfiniBand switch in turn provides connections between the leaf modules through its back plane.
 |
 |
| 1a | 1b |
Figure 1: InfiniBand network. 1a Red fiber-optic cables from compute nodes to 1b switch with 18-port leaf modulesAn All_to_All communication simultaneously sends and receives data between all parallel processes in a code. Since it is eventually not possible to have physical cable connections between all possible pairs of ports in the InfiniBand switch and its leaf modules, All_to_All commands necessarily lead to contention between the required pairwise communications. The network schematics give an impression how many cables would be needed to connect N = 9, 18, 36 nodes, respectively.
The MPI All-to-All communication command sends the block of its input array from Process
to Process
and receives it into the
block of the output array on Process
. To test the InfiniBand network, we will maximize the contention by communicating the largest block sizes possible.
We construct an algorithm that constructs a global array of vectors, each comprising
double-precision numbers, is split onto the
parallel processes. Each local array is of length
.
We maximize the contention by designing the test case to have all blocks to be of same maximum size,
parallel processes on each compute node maximizes contention on each node when al l local processes access the InfiniBand cable at the same time.
We consider two cases. The first case when we keep the gloabl memory of the array constant across all studies as increases as shown in the following test run.
| Number of nodes | N = 1 | N = 3 | N = 9 | N = 18 | N = 36 |
| Number of processes | p = 8 | p = 24 | p = 72 | p = 144 | p = 288 |
| m = 512 | 1.14 | 0.57 | 0.25 | 0.15 | 0.11 |
Table 1: Wall clock time for constant global memoryFrom the table we see that when global memory is constant, run time decreases as increases. We conclude that the All-to-Allv contention is overcome by the InfiniBand inter-connect.
Second we consider when the local memory constant, mainly across all processes. The run times can be seen in the table below.
| Number of nodes | N = 1 | N = 3 | N = 9 | N = 18 | N = 36 |
| Number of processes | p = 8 | p = 24 | p = 72 | p = 144 | p = 288 |
| m = 512 N | 0.60 | 1.64 | 2.09 | 2.28 | 2.30 |
| m = 800 N | 1.79 | 3.05 | 3.73 | 5.01 | 6.73 |
| m = 810 N | 1.80 | 2.83 | 3.30 | 5.54 | ERR |
| m = 1024 N | 85.00 | 170.62 | ERR | ERR | ERR |
Table 2: Wall clock time for constant local memoryWith local memory constant and contention on the network maximized, the run times grow with the number of processes. We can conclude that this test case creates stress on the InfiniBand network and that its performance will limit the scalability of parallel algorithms that use All_to_All communications Furthermore, for cases with larger memory requirement, we encounter excessive run times and eventually memory errors.
Clustering of Multidimensional Data Sets with Applications to Spatial Distributions of Ribosomal Proteins
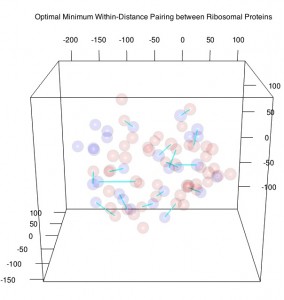
Consider ribosomal proteins, each with a three-dimensional spatial location. Proteins related to the cofactor phenotype may be randomly or non-randomly distributed within the ribosome. To investigate this question, the Mahalanobis distance is computed between each pair of protein locations, and the optimal pairing is determined by minimizing the sum of the within-pair distances. Since no single code exists that allows for the computation of Mahalanobis distances, determining the optimal pairing, and determining whether the two groups are statistically different, we created a code that allows a user to just this. The user can also compute an exact p-value for this distribution rather than rely on an approximation.
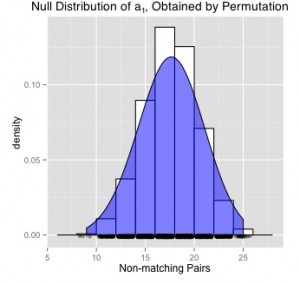
To yield conclusive evidence regarding the adjacency of phenotype and non-phenotype groups, a p-value may be determined based on the number of non-matching pairs. Intuitively, a greater number of non-matching pairs should provide evidence to similar adjacency between groups, whereas a low number of non-matching pairs should likewise lead one to expect dissimilar adjacency. Deriving the exact null distribution to the set of non-matching pairs, the null hypothesis upholding equal adjacency between groups may be rejected if A1 is small enough.
After computing the Mahalanobis distance between each protein pair within the data set, it was determined by the optimal, non-bipartite sorting algortihm that there are 17 non-matching pairs of ribosomal proteins matching phenotype to non-phenotype traits. Since the data set contains 76 elements, the maximum possible non-matching pairs is 76/2 = 38 pairs, and therefore one should notice the high proportion of non-matching pairs A1 = 17.
Therefore, a null hypothesis may be constructed claiming that the distributions of phenotype to non-phenotype positions are similarly adjacent. Computing a p-value based on the above non-matching pairs, equal to 0.54, one may reject the null hypothesis at a standard alpha level equal to 0.05. That is, one may fail to reject the null hypothesis, and conclude that the distributions comparing adjacency between phenotype and non-phenotype groups are statistically equivalent. Further, this conclusion is supported by the high proportion of non-matching pairs determined above.
 |
 |
| 4a | 4b |
Figure 4
Links
Nil Mistry, Jordan Ramsey, Benjamin Wiley, Jackie Yanchuck, Andrew Raim, Xuan Huang, Matthias K. Gobbert, Nagaraj K. Neerchal, Philip Farabaugh. Clustering of Multidimensional Data Sets with Applications to Spatial Distributions of Ribosomal Proteins. Technical Report HPCF-2013-10, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, 2013. Reprint in HPCF publications list
Nil Mistry, Jordan Ramsey, Benjamin Wiley, Jackie Yanchuck, Xuan Huang, Matthias K. Gobbert, Christopher Mineo, David Mountain.Contention of Communications in Switched Networks with Applications to Parallel Sorting. Technical Report HPCF-2013-13, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, 2013. Reprint in HPCF publications list
Poster for project one presented at the Summer Undergraduate Research Fest (SURF)
Poster for project two presented at the Summer Undergraduate Research Fest (SURF)
Click here to view Team 1’s project
Click here to view Team 2’s project
Click here to view Team 4’s project