| Team Members: | Kristen Deetz1, Nygel Foster2, Darius Leftwich 2, Chad Meyer3, Shalin Patel4 |
| Graduate Assistant: | Carlos Barajas5 |
| Faculty Mentor: | Matthias K. Gobbert5 |
| Client: | Zana Coulibaly5 |
1Department of Mathematics, Eastern University
2Department of Computer Science and Electrical Engineering , University of Maryland, Baltimore County,
3Department of Mathematics, University of Pittsburgh
4Department of Computer Science and Mathematical Science, Kean University
5Department of Mathematics and Statistics, University of Maryland, Baltimore County,
6Department of Pharmacology, University of California, Davis


About the Team
Our Team- Kristen Deetz, Nygel Foster, Darius Leftwich, Chad Meyer, and Shalin Patel- extended a partial differential equations model of calcium induced calcium release (CICR) in heart cells to show not only its electrical and chemical componenets, but also to include the mechanical contraction of the heart cell. The model was written in C an parallelized using MPI and on the HPCF cluster maya. We conducted this research at the UMBC REU Site: Interdisciplinary Program in High Perfomance Computing in the summer of 2017, with guidance from our faculty mentor, Matthias K. Gobbert and our graduate assistant Carlos Barajas.
Problem
Cardiac arrhythmias can be caused by a dysregulation of calcium dynamics in cardiomyocytes. Calcium dysregulation is not yet fully understood and is not easily predicted. However, in order to continue searching for methods to combat heart disease, it is vital that the heart and its underlying process are understood with depth. The study of the interplay between electrical excitation, calcium signaling and mechanical contraction has the potential to better our understanding of the regular functioning of the cardiomyocytes and help us understand how any dysregulation can lead to potential cardiac arrhythmias. Calcium induced calcium release (CICR) is an important part, can be modeled using a system of partial differential equations that link the electrical excitation, calcium signaling and mechanical contraction components of a cardiomyocyte.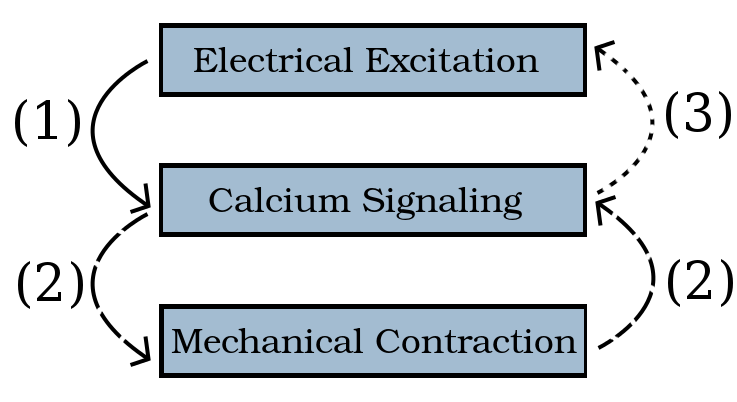
The starting point of this research, the work of 2016’s Team 5, was a model which established a ②, ③, ④ which extended 2015’s Team 5’s earlier implementation: ①.
Methodology
In our research, we built on code that was developed in C. We extended a previous model to now include the mechanical component in the cycle of a heart cell. Our new addition to the model was to include a new partial differential equation(PDE) which contracts the third cytosol buffer species. This shows us the concentration of the actin-myosin cross bridges, which influences the contraction of the heart cell. The heart cells’ process of excitation-contraction coupling, in which calcium induced calcium release is an important part, can be modeled using a system of partial differential equations. We model the contraction via the proportion of contractile proteins which have bound to calcium and changed shape as result, which generates the force required for cell contraction. Thus, performing a parameter study to determine how the interaction of electrical and calcium systems can impact the heart cells’ level of contraction.
Results
In the heart cells’ cycle, when there is an increase in voltage, calcium release units(CRUs) are opened which release calcium in that location of the cell. In the area where the calcium is released, the actin-myosin cross bridges become active, causing the heart cell to contract, therefore decreasing the shortening factor.
Iso Surface Plot
CRU Plot
Conclusions and Future Directions
We now have a working seven variable model which includes the mechanical component of the calcium induced calcium release cycle. Our research resulted in the successful implementation of the third cytosol buffer species, the inactive actin-myosin cross-bridges. The bulk of our work came through gaining an understanding of previous work, performing simulations with recent versions of the model including last year’s and the three variable case, and making necessary modifications to the previous model in C code. We incorporated new ideas and material and then simulated specific cases to get a better understanding of the behavior of the model.
We have performed several simulations, testing various parameters, in order to create the most accurate model. In particular, we manipulated the $F_{max}$ and $\kappa$ values in order to get desirable shortening factor behavior, as seen in the epsilon vs. time plots above
Because our epsilon plots are not ideal to our understanding, future research done on this topic could include further parameter studies in order to get more reasonable shortening factor behavior. Once this is established, the model can be extended to include an eighth variable.
Links
Kristen Deetz, Chad Meyer, Shalin Patel, Darius Leftwich, Nygel Foster, Carlos Barajas, Matthias K. Gobbert, and Zana Coulibaly. Coupling the Electrical and Calcium Dynamics in a Heart Cell. Technical Report HPCF-2017-15, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, 2017. (HPCF machines used: maya.). Reprint in HPCF publications list
Poster presented at the Summer Undergraduate Research Fest (SURF)
Click here to view Team 1’s project
Click here to view Team 2’s project
Click here to view Team 3’s project
Click here to view Team 4’s project
Click here to view Team 6’s project