| Team Members: | Johnlemuel Casilag1, Elizabeth Gregorio2, James Della-Giustina3, Aniebiet Jacob1 |
| Graduate Assistant: | Carlos Barajas3 |
| Faculty Mentor: | Matthias K. Gobbert3 |
| Client: | Jerimy Polf 4 |
1Department of Mathematics and Statistics, University of Maryland, Baltimore County,
2 Department of Physics, Hamline University
3School of Information Technology & Computer Science, Community College of Baltimore County
4 Department of Radiation Oncology,University of Maryland – School of Medicine
About the Team
Our team – Johnlemuel Casilag, James Della-Giustina, Elizabeth Gregorio, and Aniebiet Jacob – implemented an MPI algorithm in our code that parallelized the fast image reconstruction system. Our algorithm was written in a combination of C++ and C and utilized Matlab for imaging purposes, and was fully parallelized using MPI on the HPCF cluster maya. We conducted our research in the summer of 2017 at the UMBC REU Site: Interdisciplinary Program in High Performance Computing, with guidance and assistance from our faculty mentor Matthias K. Gobbert and our graduate assistant Carlos Barajas. The image on the right is Dr. Gobbert, our Team, and Carlos all at the Proton Beam Therapy Center with the client Dr. Jerimy Polf.
Problem
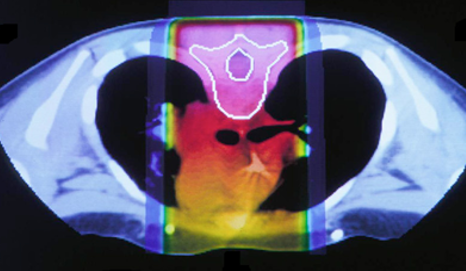
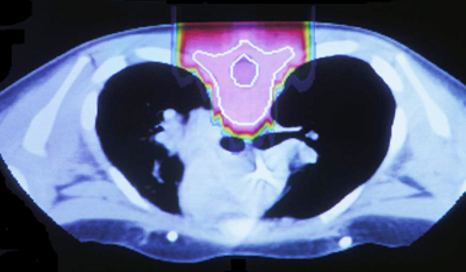
The idea to utilize proton beams as radiotherapy stems from the realization that while x-ray’s deliver a lethal dose of radiation throughout the patient, proton beams will reach their highest dose just before they stop, at what is called the Bragg peak. This comparison can be seen in the images below. The left image shows traditional x-ray therapy, in which the beam goes across the whole body. The right image shows proton beam therapy, where the rays stop at a fixed depth into the body. This makes proton beams are preferable from a clinical standpoint because they have the potential to reduce the radiation dose delivered to healthy tissue surrounding the tumor.
Although there is great potential to reduce healthy tissue exposure through radiation when using proton beam radiation, this potential cannot always be fully taken advantage of because of uncertainties. These uncertainities stem from the inability for the physician to see clearly where the dose of radiation from the proton beam is deposited, which is why these proton beams must be imaged.
This imaging is possible because of the interaction between the proton beam and the elements that compose our cells. This interaction releases gamma rays, therefore if the origin of the gamma ray can be traced back, then the doctor can see where the proton beam is delivering its dose. This is called prompt gamma ray imaging and offers doctors the ability to make adjustments to treatment in real time. However, in order for the proton beam to hit precisely where the physician intends, it is necessary for the patient to lie entirely still during treatment. This same requirement is true for imaging. Therefore, the run time of the software needs to be as fast as possible, which is what our team tried to accomplish.
Methodology
Building upon the already developed C++ code by the clients, our team integrated MPI into the code. In addition, in order to most efficiently map our results and implement our changes we translated a few of the C++ functions into C code and in addition created a new program for mapping the gramma emission into a hisogram using Matlab code.
Results
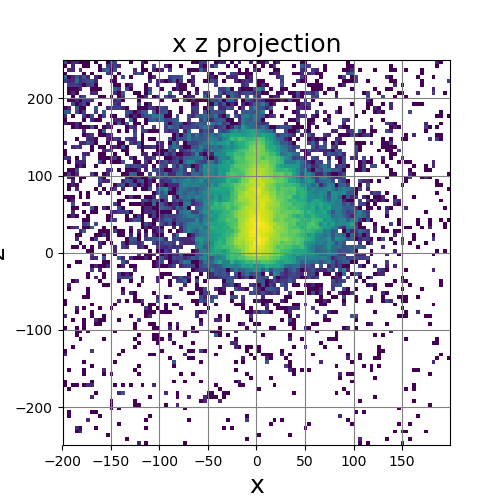
The following images show the results of a cross-section of the reconstructed images after 100, 300, and 600 iterations, respectively. The first set of images were computed by the original algorithm and post-processed with its Python code. The second set of images were computed by our MPI code and using our Matlab code for post-processing.
OpenMP – Serial
 |
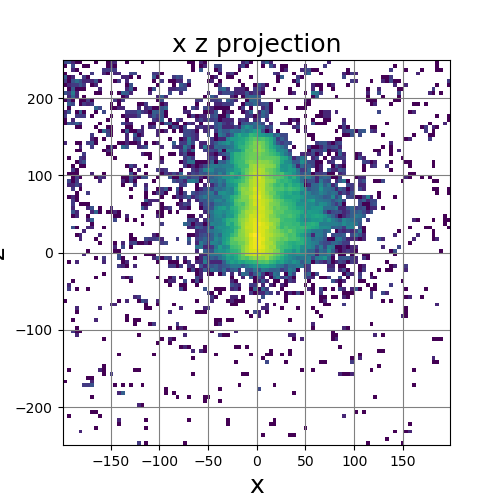 |
 |
| 100 Iterations | 300 Iterations | 600 Iterations |
MPI – Serial
 |
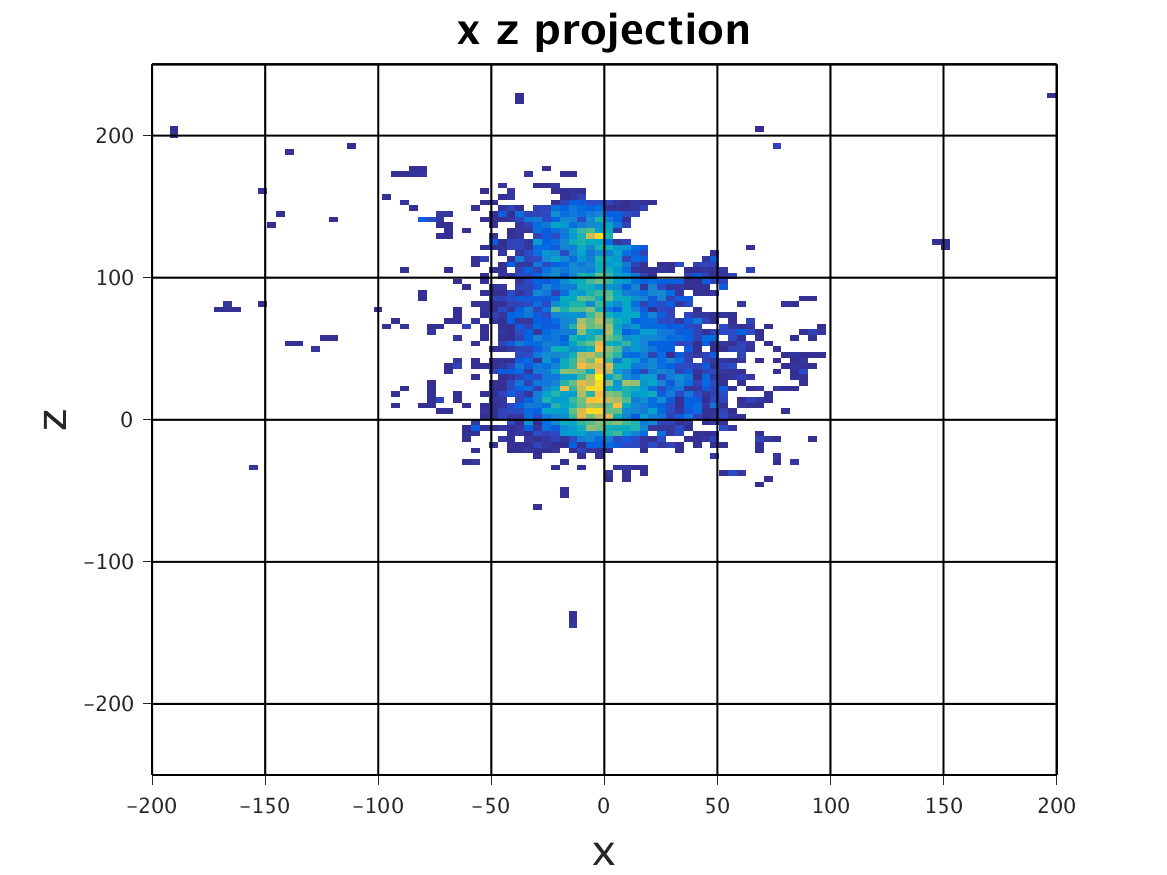 |
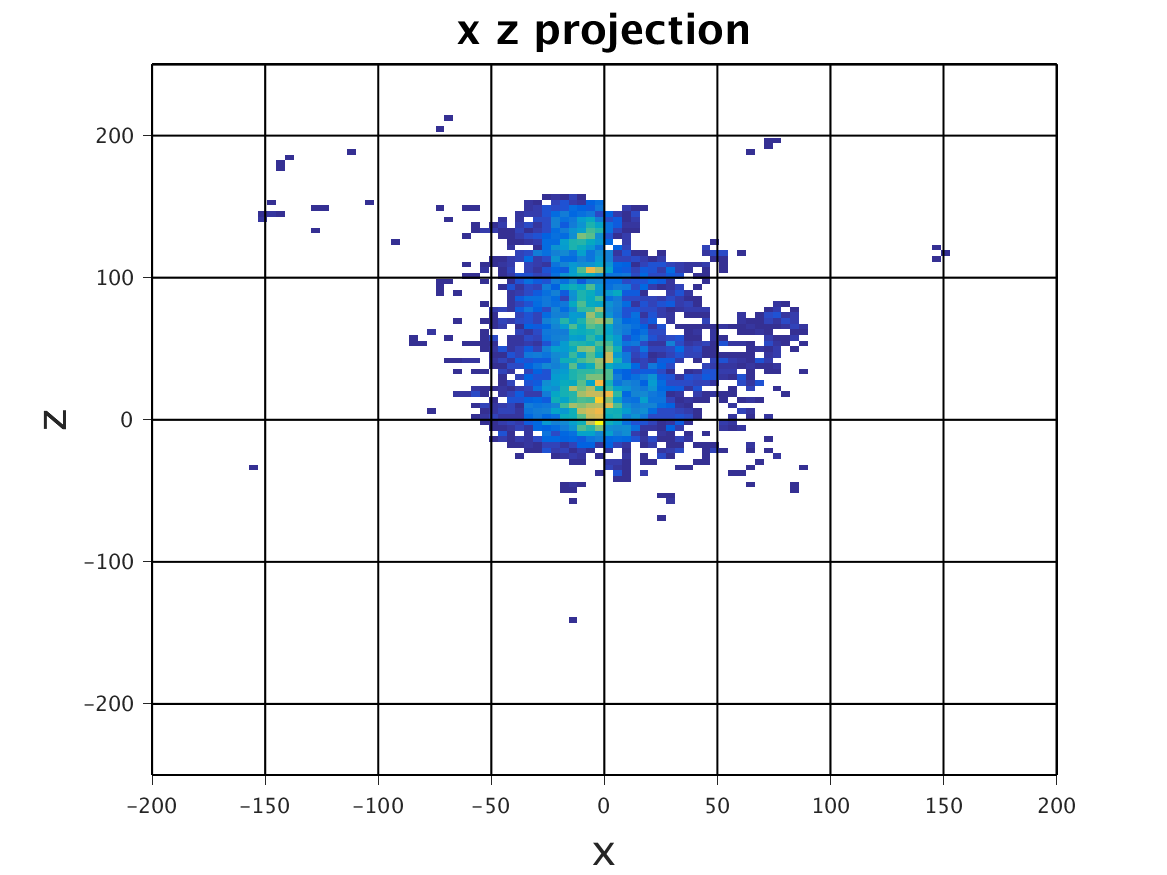 |
| 100 Iterations | 300 Iterations | 600 Iterations |
The images have a different appearance resulting from the use of different plotting techniques, but they show convergence to the same position of the sample. This confirms the correctness of the code. The images shown for the MPI code come from the second version of MPI code, termed modified MPI algorithm, in which the algorithm is actually changed by communicating less frequently, which increases parallel efficiency.
The following table are the timing results that the OpenMP multithreading algorithm, the original MPI algorithm, and the modified MPI algorithm code produced. This table presents the observed wall clock time in seconds for reconstruction of image from 100,000 cones with 102-by-102-by-126 bins in the 3-D histogram using 600 iterations with each algorithm using 1, 2, 4, etc. many computational cores. In the case of OpenMP, the numbers of threads are limited to the cores in one compute node with two 8-core CPUs, while for MPI, two and four nodes can be pooled to use 32 and 64, respectively, processes.
| Computational cores: | 1 | 2 | 4 | 8 | 16 | 32 | 64 |
| OpenMP multi-threading (s) | 1885 | 889 | 344 | 188 | 105 | N/A | N/A |
| Original MPI algorithm (s) | 1592 | 546 | 372 | 354 | 511 | 477 | 430 |
| Modified MPI algorithm (s) | 985 | 485 | 277 | 184 | 148 | 392 | 83 |
The multi-threaded algorithm using OpenMP exhibits good speedup since it runs faster with multiple threads than using one thread, however the OpenMP algorithm is limited to one node therefore it cannot be parallelized beyond 16 threads and thus limited to 105 seconds as fastest result for this test case. Because of this, we decided to change the OpenMP implementation of the code to an MPI algorithm implementation. However, though the initial MPI algorithm computes reconstruction of image to same quality as the OpenMP version, it did not show good performance, although it can scale to multiple nodes. Ultimately, we modified this MPI algorithm, and the modified MPI algorithm shows much better performance than both the OpenMP algorithm and the initial MPI algorithm. The fastest time that was presented from this code was 83 seconds for 64 processes, using 16 cores on 4 nodes, which is something that the shared-memory OpenMP algorithm is not able to do.
Conclusions and Future Directions
The original algorithm and code provided by Dr. Polf and Dr. Mackin implemented solely OpenMP, placing a ceiling on performance. With the inability to utilize distributed memory, we were bound to the performance of one node and its associated memory. By implementing MPI into our code, we were able to reduce the best performance of the original OpenMP code from 105 seconds down to 85 seconds. While this may not seem like a huge improvement, MPI’s distributed memory and multiple-processing power shows the potential to scale these run times down even further by using more nodes and in conjunction with OpenMP for each MPI process. The key potential for additional speedup lies in optimizing the code more and in using hybrid MPI+OpenMP code on cutting-edge computer hardware such as multi-core CPUs with more cores or the many-core Intel Xeon Phi with 64 cores. This in the end would be essential to making sure that this program could be utilized in a clinical setting.
Links
Elizabeth Gregorio, James Della-Giustina, Aniebet Jacob, Johnlemuel Casilag, Carlos Barajas, Matthias K. Gobbert, Dennis S. Mackin and Jerimy Polf. Development of Fast Reconstruction Techniques for Prompt Gamma Imaging During Proton Radiotherapy. HPCF-2017-16, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, 2017. Reprint in HPCF publications list
Poster presented at the Summer Undergraduate Research Fest (SURF)
Click here to view Team 1’s project
Click here to view Team 2’s project
Click here to view Team 3’s project
Click here to view Team 4’s project
Click here to view Team 5’s project