| Team Members: | Ely Biggs1, Tessa Helble2, George Jeffreys3, and Amit Nayak4 |
| Graduate Assistant: | Elias Al-Najjar2 |
| Faculty Mentor: | Kofi P. Adragni2 |
| Client: | Andrew Raim5 |
1Department of Applied Mathematics, Wentworth Institute of Technology,
2Department of Mathematics and Statistics, University of Maryland, Baltimore County,
3Department of Mathematics and Statistics, Rutgers University,
4Department of Mathematics, George Washington University,
5United States Census Bureau
About the Team
Our team, Ely Biggs, Tessa Helble, George Jeffreys, and Amit Nayak participated in the Interdisciplinary Program in High Performance Computing located in the Department of Mathematics and Statistics at UMBC under the guidance of Dr. Kofi Adragni, research assistant Dr. Elias Al-Najjar of the Mathematics and Statistics Department at UMBC and client Dr. Andrew M. Raim of the Census Bureau. We undertook the challenge of parallelizing the code that they wrote for their new statistical methodology called MADE (Minimum Average Deviance Estimation), as well as analyzing its performance on the cluster maya.
The Master Address File (MAF)
The United States Census Bureau maintains a massive list of information about all habitable addresses in the U.S. called the Master Address File (MAF). Verifying all of the information in the MAF via a process called Address Canvassing (AdCan) was the second most expensive part of the 2010 census. Address canvassing involves recording all addresses that must to be added to the MAF (new habitable addresses) and all addresses that must be deleted (no longer habitable).
MADE is a statistical methodology being developed by Kofi Adragni, Andrew Raim, and Elias Al-Najjar for the purpose of analyzing and making predictions using Poisson counts of deletions and additions to the MAF with the goal of using these predictions to reduce the cost of future AdCans.
The difficulty in developing a methodology is the size of the data set, as it can have hundreds of millions of observations and thousands of variables for each observation, thus having incredibly high dimensions. MADE embeds a sufficient dimension reduction procedure, along with a local linear regression.
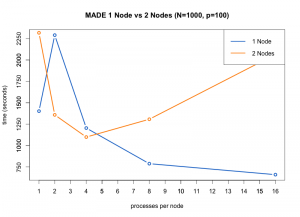
Run time of the MADE functions for N = 1000, p = 100.
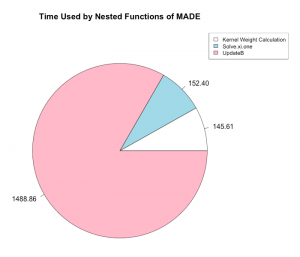
We also measured the times of all of the important functions nested inside of MADE.
Conclusions:
We were able to find that the optimal configuration of nodes and processes per node to run the MADE function in parallel on the High Performance Computer (HPC) maya at UMBC was one node and 16 processes per node. We were also able to determine that the limit of the largest dimensions of the input that MADE could handle was between 5000 by 100 and 10000 by 100, that the code takes the most time when optimizing over the Stiefel manifold, and that parallelization improved the performance of the overall code with a speedup of double that of the unparallelized code.
Links
Ely Biggs, Tessa Helble, George Jeffreys, Amit Nayak, Elias Al-Najjar, Kofi P. Adragni, and Andrew Raim. Numerical evaluation of minimum average deviance estimation in ultra high dimensional Poisson regression. Technical Report HPCF-2015-21, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, 2015. (HPCF machines used: maya.). Reprint in HPCF publications list
Poster presented at the Summer Undergraduate Research Fest (SURF)
Click here to view Team 2’s project
Click here to view Team 3’s project
Click here to view Team 4’s project
Click here to view Team 5’s project
Click here to view Team 6’s project
Click here to view Team 7’s project
Click here to view Team 8’s project
