| Team Members: | Sidafa Conde1, Teresa Lebair2, Christopher Raastad3, and Virginia Smith4 |
| Graduate Assistants: | Kyle Stern5 and David Trott5 |
| Faculty Mentor: | Matthias Gobbert5 |
| Clients: | Bradford Peercy5 and Arthur Sherman6 |
1University of Massachusetts Dartmouth, North Dartmouth, MA
2Saint Vincent College, Latrobe, PA
3University of Washington, Seattle, WA
4University of Virginia, Charlottesville, VA
5University of Maryland, Baltimore County, Baltimore, MD
6National Institutes of Health, Bethesda, MD

Team 1, from left to right: Christopher Raastad, Virginia Smith, Teresa Lebair, Sidafa Conde
In the REU Site: Interdisciplinary Program in High Performance Computing in the Department of Mathematics and Statistics at the University of Maryland, Baltimore County (UMBC), during the summer of 2010, our team of four students Sidafa Conde, Teresa Lebair, Christopher Raastad, and Virginia Smith, researched ways of enabling more effective ways of implementing physiologically representative models of pancreatic beta cell islets, under the guidance of Dr. Matthias Gobbert. This project was proposed to us by Dr. Bradford Peercy and Dr. Arthur Sherman.
Diabetes is a disease distinguished by an abnormally high concentration of glucose in the blood stream. Approximatly 23.6 million children and adults in the United States, 7.8% of the population, suffer from the desease (American Diabetes Association). The condition results either from defects in insulin production within the pancreas or from defects in functionality as the insulin interacts with cells throughout the body. Within the endocrine system of the pancreas lie clusters of cells called the islets of Langerhans. Each islet is composed of four cell types, the most prevalent of which is the beta cell.
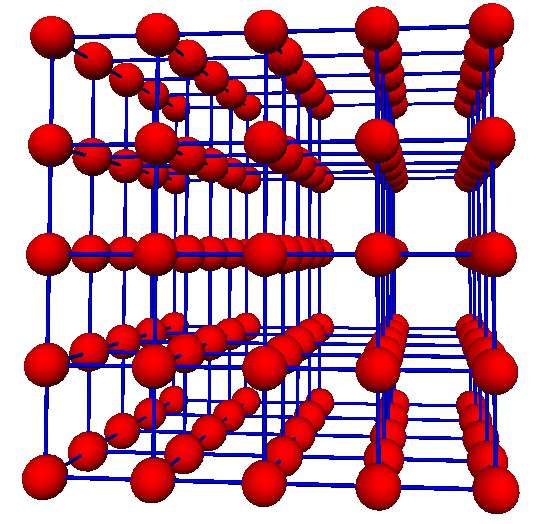
The individual beta cells within an islet of Langerhans are believed to be coupled, that is joined together by proteins, called gap junctions. These gap junctions create pores that allow the passage of ionic and small molecular material between two adjacent cells. Therefore, our model takes into account nearest neighbor coupling between cells and for ease we take these cells to exist on a cubic lattice of N-by-N-by-N cells, as shown in this schematic:
A recently proposed dual oscillator model combines electrical and metabolic mechanisms to explain the pulsing activity of the insulin secretion from beta cells. This model is given by a set of seven ordinary differential equations for each cell. Individual beta cells can be classified by their electrical patterns as either slow or fast bursting cells. The insulin secretion of each of these cell types follows the electrical oscillatory pattern: a fast cell secretes insulin at a relatively constant elevated level having smoothed out the relatively fast electrical oscillations, while a slow cell secretes insulin with a period of roughly 4 to 6 minutes. The islets of Langerhans contain both slow and fast bursting beta cells, although the proportion and distribution of slow and fast cells within the islets is currently unknown.
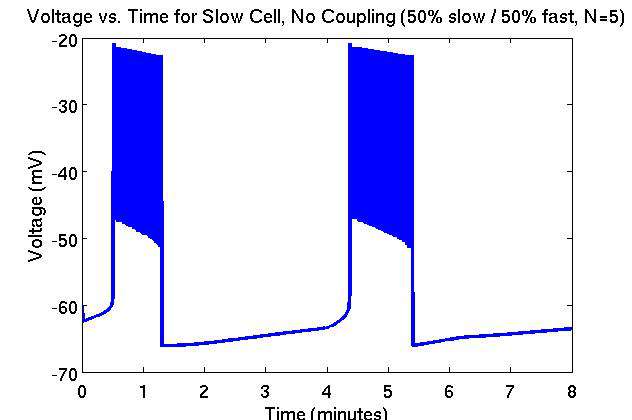
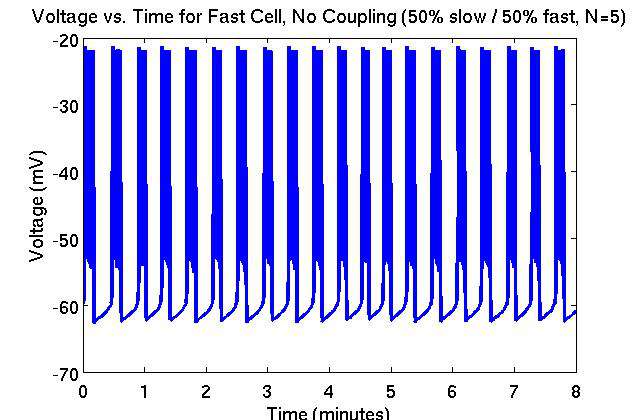
The figures show the results of a simulation with 5x5x5 cells, in an islet containing half slow and half fast cells, uncoupled. Figure 1 and 2 show the trace of voltage over time in a slow and fast bursting beta-cell, respectively. The slow cell exhibits an average burst period of 3.98 minutes, whereas the fast cell exhibits an average burst period of 0.39 minutes (24 seconds).
In the eight weeks of the program, we accomplished the following:
- The implementation of the computational model for an islet of beta cell was improved in efficiency: Key modification’s to Matlab’s ODE solver ode15s reduced the memory usage dramatically and enabled simulations up to N=10 instead of 2 or 3. The research was truly interdisciplinary and close interaction with the application researchers led to a more efficient design of the simulations by reducting the number of trials for proportions of fast-slow cells to no more than 11 for larger N. Simulations with N=10 still take 18 hours, but using 11 nodes of the cluster tara in the UMBC High Performance Computing Facility simultaneously accomplishes the complete desired simulation for the largest desired N=10 within that time.
- The implementation of the computational model for an islet of beta cell was also extended in functionality: We extended the implementation of the model by implementing a beta cell islet with random coupling strengths. By running simulations with varying coupling strength, we simulated the impact of coupling within the islets. In particular, we ran simulations for different coupling strengths on a variety of islets, each with a different combination of slow and fast cells.
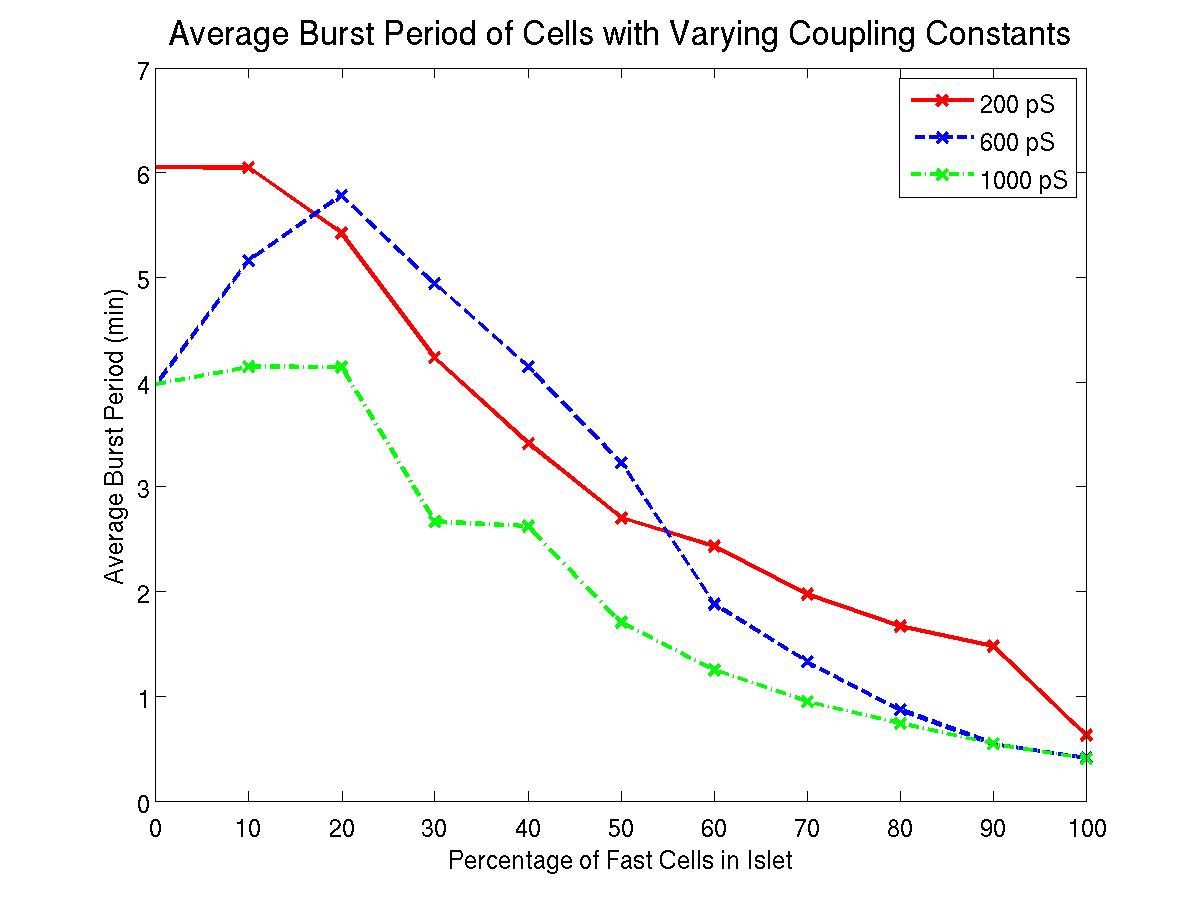
The simulations of the beta cell islet model led to a potentially transformative discovery: The figure displays the average burst period of cells in an islet with 5x5x5 cells, with varying proportions of fast and slow cells. The plot is shown for three different strengths of coupling: 200, 600, and 1000 pS. In the extreme cases of weak and of strong coupling, an increase of the proportion of fast cells lessens the average burst period. However, for intermediate values of the coupling strength, a small increase in the number of fast cells acts by first increasing the burst period, actually slowing down the oscillations, before falling into the pattern of reducing the burst period with larger proportions of fast cells again. This result is counter-intuitive and warrants further study.
Links
Sidafa Conde, Teresa Lebair, Christopher Raastad, Virginia Smith, Kyle Stern, David Trott, Matthias K. Gobbert, Bradford E. Peercy, and Arthur Sherman. Enabling Physiologically Representative Simulations of Pancreatic Beta Cells. Technical Report HPCF-2010-21, UMBC High Performance Computing Facility, University of Maryland, Baltimore County, 2010. Reprint in HPCF publications list